Classification analysis assigns an observation to a group based on its measured variables, using patterns from known group data.
Imagine sorting fruit into baskets: you measure features (e.g., weight, color), compare them to known apples and oranges, and pick the closest match.
A new observation vector is compared to previous observartions to predict its group, often via a discriminant score.
Purpose of classification
Classification predicts group membership for unknown observations, using prior samples. It’s valuable for:
- Prediction (e.g., student success, mental illness category).
- Decision-making (e.g., identifying “killer” bees).
- Allocation (e.g., matching trainees to programs).
Unlike discriminant analysis, it focuses on allocation, not just separation.
Classification procedures
- Classifying Observations into Two Groups
- Classifying Observations into Several Groups
- Nonparametric Classification Procedures
Estimating misclassification rates
-
Use training data to define the classification rule.
-
Apply the rule to every observation in the training set to predict its group.
-
For each , check if the predicted group matches its actual group.
-
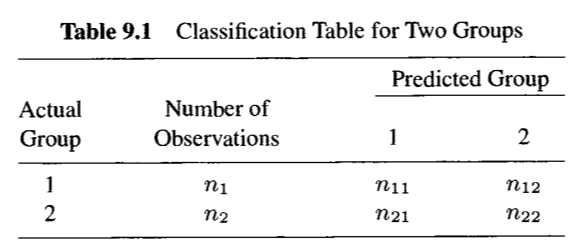
Count Misclassifications: Use a classification table (such as Table 9.1)
For 2 groups:
- : Correctly classified into
- : Misclassified into
- : Misclassified into
- : Correctly classified into .
For groups, sum all off-diagonal counts in the classification table (e.g., ).
-
Compute Rate (Formula 9.16):
\text{Apparent Error Rate} &= \frac{\text{Total Misclassifications}}{\text{Total Observations}}\\ &= \frac{n_{12} + n_{21}}{n_{1} + n_{2}}\\ &= \frac{n_{12} + n_{21}}{n_{11} + n_{12} + n_{21} + n_{22}}\\ \end{align}$$
Classification table example:
Python example
For classification into two groups, see example on Python example
The example below demonstrates 2 classifications into 3 groups, one assuming equal covariance, the other assuming unequal covariance.
import numpy as np
# Football data means (Example 9.3.1, 6 variables simplified to 2 for brevity)
y_bar_1 = np.array([15.2, 58.9]) # Group 1
y_bar_2 = np.array([15.4, 57.4]) # Group 2
y_bar_3 = np.array([15.6, 57.8]) # Group 3
means = [y_bar_1, y_bar_2, y_bar_3]
# Simplified covariance matrices
S_pl = np.array([[2.5, 0.8], [0.8, 3.0]]) # Pooled (equal covariance)
S_1 = np.array([[2.0, 0.5], [0.5, 2.5]]) # Group 1
S_2 = np.array([[2.8, 0.9], [0.9, 3.2]]) # Group 2
S_3 = np.array([[2.2, 0.7], [0.7, 2.8]]) # Group 3
covs = [S_1, S_2, S_3]
# New observation (first in Group 1)
y_new = np.array([13.5, 57.2])
# Equal Covariance (Formula 9.11)
S_pl_inv = np.linalg.inv(S_pl)
L = []
for i, y_bar in enumerate(means):
term1 = y_bar @ S_pl_inv @ y_new
term2 = 0.5 * y_bar @ S_pl_inv @ y_bar
L_i = term1 - term2 # L_i(y)
L.append(L_i)
group_linear = np.argmax(L) + 1
# Unequal Covariance (Formula 9.15, assume equal priors p_i = 1/3)
Q = []
for i, (y_bar, S_i) in enumerate(zip(means, covs)):
S_i_inv = np.linalg.inv(S_i)
diff = y_new - y_bar
term1 = np.log(1/3) # ln p_i
term2 = 0.5 * np.log(np.linalg.det(S_i))
term3 = 0.5 * diff @ S_i_inv @ diff
Q_i = term1 - term2 - term3 # Q_i(y)
Q.append(Q_i)
group_quadratic = np.argmax(Q) + 1
print("Linear scores:", list(map(int, L)))
print("Predicted group (linear):", group_linear)
print("Quadratic scores:", list(map(int, Q)))
print("Predicted group (quadratic):", group_quadratic)Linear scores: [545, 545, 545]
Predicted group (linear): 2
Quadratic scores: [-2, -2, -2]
Predicted group (quadratic): 2