When data deviates from normality, nonparametric methods classify observations without assuming a specific distribution.
This note will describe 4 nonparametric classification procecures.
Each uses sample data directly to assign to one of groups.
Multinomial classification
Treats variables as categorical (e.g., counts or discrete levels), comparing observed frequencies to expected ones per group.
For an observation with frequency in group :
- Assign to if (Formula 9.17, two groups), else .
- For , assign to group maximizing (generalized rule).
Steps:
- Categorize Data: Convert continuous into discrete levels (e.g., bins) or use naturally categorical data.
- Count Frequencies: For each category in , count occurrences in training data for group .
- Estimate Priors: Set prior probability (e.g., or equal if unspecified).
- Compute Ratios (2 Groups): For each category , calculate and compare to .
- Classify: If ratio > threshold, assign to ; else . For , compute for all and pick max.
Classification based on density estimators
Estimates the probability density for each group using a kernel (e.g., normal) and assigns to the group with the highest density.
Steps
-
Compute kernel density estimate (9.23):
where
- : Kernel (e.g., normal)
- : Smoothing parameter (e.g., from Table 9.8)
- : Amount of variables
- : Sample size
-
Assign to group which has the maximum (9.28), where is the -th prior probability.
Nearest neighbor classificaton rule
Assigns to the group most common among its closest observations, based on distance.
Steps:
-
Compute distances of to other points using the distance function:
-
Assign to group with highest count among nearest neighbors. For 2 groups, assign to if:
Or for further refinement, use prior probabilities:
For groups, assign the observation to the group that has the highest Where:
- number of observations from among the nearest neighbors of the observation in question.
We suggest choosing nearing .
In practice, one could test several values of , and use one with the best error rate.
When data deviates from normality, nonparametric methods classify observations without assuming a specific distribution.
This note will describe 4 nonparametric classification procecures.
Each uses sample data directly to assign to one of groups.
Multinomial classification
Treats variables as categorical (e.g., counts or discrete levels), comparing observed frequencies to expected ones per group.
For an observation with frequency in group :
- Assign to if (Formula 9.17, two groups), else .
- For , assign to group maximizing (generalized rule).
Steps:
- Categorize Data: Convert continuous into discrete levels (e.g., bins) or use naturally categorical data.
- Count Frequencies: For each category in , count occurrences in training data for group .
- Estimate Priors: Set prior probability (e.g., or equal if unspecified).
- Compute Ratios (2 Groups): For each category , calculate and compare to .
- Classify: If ratio > threshold, assign to ; else . For , compute for all and pick max.
Classification based on density estimators
Estimates the probability density for each group using a kernel (e.g., normal) and assigns to the group with the highest density.
Steps
-
Compute kernel density estimate (9.23):
where
- : Kernel (e.g., normal)
- : Smoothing parameter (e.g., from Table 9.8)
- : Amount of variables
- : Sample size
-
Assign to group which has the maximum (9.28), where is the -th prior probability.
Nearest neighbor classificaton rule
Assigns to the group most common among its closest observations, based on distance.
Steps:
-
Compute distances of to other points using the distance function:
-
Assign to group with highest count among nearest neighbors. For 2 groups, assign to if:
Or for further refinement, use prior probabilities:
For groups, assign the observation to the group that has the highest Where:
- number of observations from among the nearest neighbors of the observation in question.
We suggest choosing nearing .
In practice, one could test several values of , and use one with the best error rate.
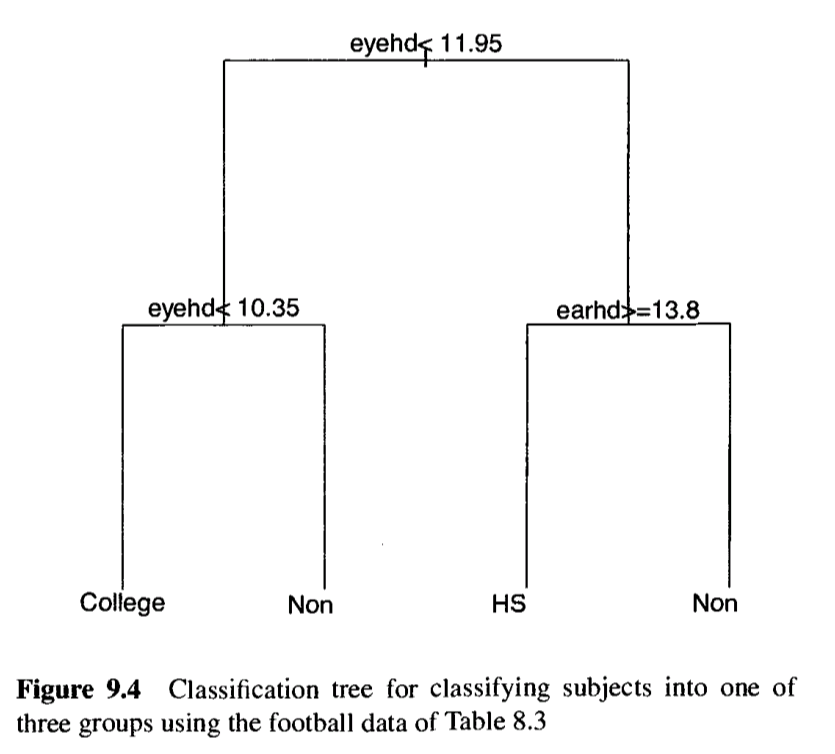
Classification Trees
Builds a decision tree by recursively splitting data into nodes based on predictor variables, assigning to the group most common in its terminal node.
Steps:
-
Start at root node: Place all observations (training data) in one group (root node).
-
Choose split: For each predictor variable , test all possible cutoffs to split the node into two child nodes ( and ):
- Compute impurity (Gini index, 9.32) for the parent node , where (9.35).
- Compute impurity for child nodes and .
- Calculate change in impurity: (9.36), where .
- Select the variable and cutoff maximizing (best split).
-
Repeat recursively: Apply step 2 to each child node (e.g., , ) until a stopping rule (e.g., cross-validation) determines the optimal tree size. Nodes that stop splitting are terminal nodes.
-
Assign groups: For each terminal node, assign the group with the highest count.
-
Classify : Traverse the tree with ‘s values:
- Start at the root, follow splits (e.g., if eyehd < 11.95, go left).
- Reach a terminal node and assign to its group.
Python example
import numpy as np
from scipy.stats import multivariate_normal
# Simplified football data (3 groups, 2 variables: e.g., speed, strength)
G1 = np.array([[15.2, 58.9], [14.8, 59.1], [15.0, 58.7], [15.4, 59.0], [14.9, 58.8]])
G2 = np.array([[15.4, 57.4], [15.6, 57.2], [15.3, 57.6], [15.5, 57.3], [15.2, 57.5]])
G3 = np.array([[15.6, 57.8], [15.8, 57.6], [15.7, 57.9], [15.5, 57.7], [15.9, 57.8]])
X = np.vstack([G1, G2, G3]) # All training data
y = np.array([1]*5 + [2]*5 + [3]*5) # Labels: 1, 2, 3
n_groups = 3
n_per_group = 5
new_y = np.array([13.5, 57.2]) # New observation
# --- Multinomial Classification ---
# Bin into 3 categories per variable (low, med, high)
bins = [np.percentile(X[:, i], [0, 33, 66, 100]) for i in range(2)]
y_cat = [np.searchsorted(bins[i][1:-1], new_y[i], side='right') for i in range(2)]
X_cat = np.array([np.searchsorted(bins[i][1:-1], X[:, i], side='right') for i in range(2)]).T
cat_idx = X_cat[:, 0] * 3 + X_cat[:, 1] # Index for 2D categories
new_idx = y_cat[0] * 3 + y_cat[1]
# Counts for each group in the new category (with small smoothing to avoid zeros)
q_hi = [np.sum((cat_idx == new_idx) & (y == h)) + 0.01 for h in range(1, 4)]
p_h = [1/3] * 3 # Equal priors
scores_multinomial = [p_h[i] * q_hi[i] for i in range(3)]
group_multinomial = np.argmax(scores_multinomial) + 1
# --- Density Estimation ---
h = 2.0 # Bandwidth from Table 9.8
p = 2 # Dimensions
kernel_const = (2 * np.pi)**(-p/2) # Normal kernel constant
f_h = []
for h_idx in range(n_groups):
group_data = X[h_idx * n_per_group:(h_idx + 1) * n_per_group]
kernel_sum = 0
for x in group_data:
u = (new_y - x) / h
kernel_sum += kernel_const * np.exp(-0.5 * np.sum(u**2))
density = kernel_sum / (n_per_group * h**p)
f_h.append(density)
scores_density = [p_h[i] * f_h[i] for i in range(n_groups)]
group_density = np.argmax(scores_density) + 1
# --- Nearest Neighbor (k=5) ---
distances = np.sqrt(np.sum((X - new_y)**2, axis=1)) # Euclidean distance
k = 5
nearest_idx = np.argsort(distances)[:k] # Indices of k smallest distances
nearest_labels = y[nearest_idx]
counts = np.zeros(n_groups + 1, dtype=int) # Manual bincount
for label in nearest_labels:
counts[label] += 1
group_nn = np.argmax(counts[1:]) + 1 # Ignore index 0
# --- Classification Tree ---
# Fixed tree with proper branching
def classify_tree(x, X, y):
if x[0] < 15.5: # First split
if x[1] < 58.0: # Second split for left branch
region = (X[:, 0] < 15.5) & (X[:, 1] < 58.0)
else:
region = (X[:, 0] < 15.5) & (X[:, 1] >= 58.0)
else: # Right branch of first split
if x[1] < 58.0: # Second split for right branch
region = (X[:, 0] >= 15.5) & (X[:, 1] < 58.0)
else:
region = (X[:, 0] >= 15.5) & (X[:, 1] >= 58.0)
region_labels = y[region]
if len(region_labels) == 0: # Default to most common group
return np.argmax(np.bincount(y)[1:]) + 1
group_counts = np.bincount(region_labels, minlength=n_groups+1)
return np.argmax(group_counts[1:]) + 1
group_tree = classify_tree(new_y, X, y)
# Results - format as floats to preserve small values
print("Multinomial scores:", [f"{score:.6e}" for score in scores_multinomial])
print("Predicted group (multinomial):", group_multinomial)
print("Density scores:", [f"{score:.6e}" for score in scores_density])
print("Predicted group (density):", group_density)
print("Nearest neighbor counts for groups 1,2,3 (k=5):", counts[1:])
print("Predicted group (nearest neighbor):", group_nn)
print("Predicted group (tree):", group_tree)Multinomial scores: ['3.333333e-03', '3.366667e-01', '3.333333e-03']
Predicted group (multinomial): 2
Density scores: ['6.801202e-03', '8.378014e-03', '6.959560e-03']
Predicted group (density): 2
Nearest neighbor counts for groups 1,2,3 (k=5): [0 4 1]
Predicted group (nearest neighbor): 2
Predicted group (tree): 2